Metrics Summary CSV
File: [sample_name]_Metrics_Summary.csv
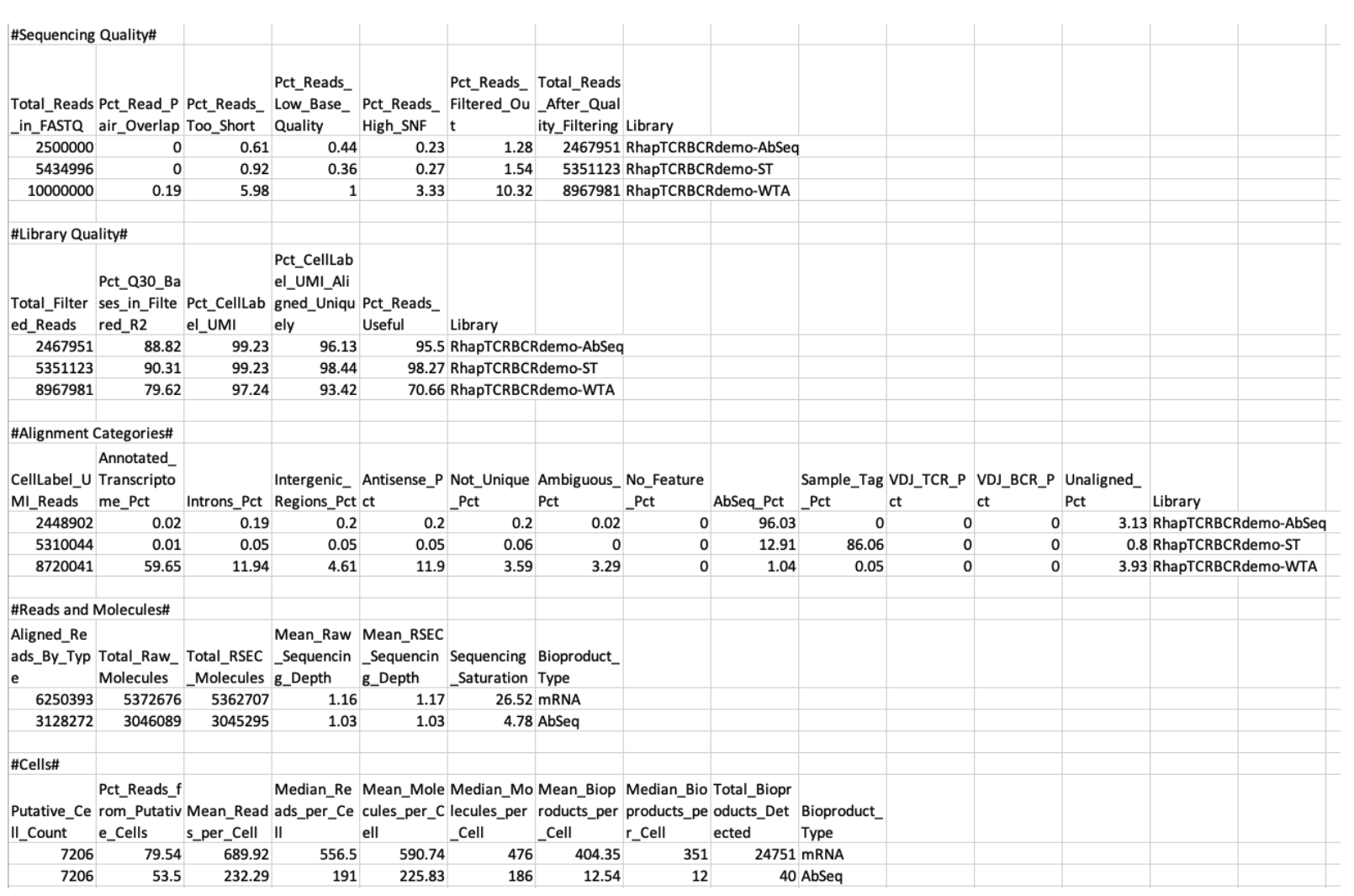
The Metrics summary provides statistics on sequencing, molecules, cells, and bioproducts.
Sample Tag, VDJ and AbSeq metrics display only when they are used in an experiment.
Metrics summary output sections and metrics
Sequencing Quality
| Metric | Definition | Major contributing factors |
|---|---|---|
| Total_Reads_in_FASTQ | Number of read pairs in the input FASTQ files | Sequencing amount |
| Pct_Read_Pair_Overlap | Percentage of read pairs overlapped with each other | Library quality |
| Pct_Reads_Too_Short | Percentage of read pairs filtered out due to length | Sequencing quality |
| Pct_Reads_Low_Base_Quality | Percentage of reads filtered out due to average base quality score <20 | Sequencing quality |
| Pct_Reads_High_SNF | Percentage of read pairs filtered out due to single nucleotide frequency ≥55% for R1 or ≥80% for R2 | Sequencing quality |
| Pct_Reads_Filtered_Out | Percentage of reads removed by the combination of length, quality, and SNF filters | Sequencing quality |
| Library | Name of library | Name of library |
Library Quality
| Metric | Definition | Major contributing factors |
|---|---|---|
| Total_Filtered_Reads | Number of read pairs after length, quality, and SNF filtering | Sequencing amount Sequencing quality Library quality |
| Pct_Q30_Bases_in_Filtered_R2 | Percentage of R2 bases with quality score >30, averaged across all read pairs retained after quality filtering | Sequencing quality |
| Pct_CellLabel_UMI | Percentage of read pairs containing a valid cell label and UMI | Sequencing quality Library quality |
| Pct_CellLabel_UMI_Aligned_Uniquely | Percentage of read pairs containing a valid cell label and UMI that aligned uniquely | Sequencing quality Library quality |
| Pct_Reads_Useful | Percentage of read pairs containing a valid cell label and UMI that aligned uniquely to a valid bioproduct | Sequencing quality Library quality |
| Library | Name of library | Name of library |
Alignment Categories
| Metric | Definition | Major contributing factors |
|---|---|---|
| Total_CellLabel_UMI_Reads | Number of read pairs containing a valid cell label and UMI | Sequencing quality Library quality |
| Annotated_Transcriptome_Pct | Percentage of cellular reads with read 2 aligned uniquely to gene present in the transcriptome (WTA) or mRNA panel (Targeted) | Sequencing quality Library quality Cell type |
| Introns_Pct | Percentage of cellular reads with read 2 aligned uniquely to an intronic region of a gene | Sequencing quality Library quality Cell type |
| Intergenic_Regions_Pct | Percentage of cellular reads with read 2 aligned uniquely to an intergenic region | Sequencing quality Library quality Cell type |
| Antisense_Pct | Percentage of cellular reads with read 2 aligned uniquely to an antisense strand | Sequencing quality Library quality Cell type |
| Not_Unique_Pct | Percentage of cellular reads with read 2 not uniquely aligned | Sequencing quality Library quality Cell type |
| Ambiguous_Pct | Percentage of cellular reads with read 2 aligned to region with ambiguous annotation | Sequencing quality Library quality Cell type |
| No_Feature_Pct | Percentage of cellular reads with read 2 aligned to non-annotated region (WTA) or targeted mRNA reads that are filtered out due to an invalid alignment | Sequencing quality Library quality Cell type |
| AbSeq_Pct | Percentage of cellular reads with read 2 aligned to AbSeq reference | Sequencing quality Library quality |
| Sample_Tag_Pct | Percentage of cellular reads with read 2 aligned to Sample Tag | Sequencing quality Library quality |
| VDJ_TCR_Pct | Percentage of cellular reads with read 2 aligned to TCR gene segments | Sequencing quality Library quality |
| VDJ_BCR_Pct | Percentage of cellular reads with read 2 aligned to BCR gene segments | Sequencing quality Library quality |
| Unaligned_Pct | Percentage of cellular reads with read 2 unaligned to reference | Sequencing quality Library quality |
| Library | Name of library | Name of library |
Reads and Molecules
| Metric | Definition | Major contributing factors |
|---|---|---|
| Aligned_Reads_By_Type | Number of filtered read pairs aligned to bioproduct type | Sequencing quality Library quality |
| Total_Raw_Molecules | Total number of molecules as defined by the unique combination of cell label, bioproduct identity, and UMI | Sequencing depth Library quality |
| Total_RSEC_Molecules | Total number of molecules detected after the RSEC molecular identifier adjustment algorithm | Sequencing depth Library quality |
| Mean_Raw_Sequencing_Depth | Average number of read pairs per molecule before molecular identifier adjustment algorithms | Sequencing depth |
| Mean_RSEC_Sequencing_Depth | Average number of read pairs per molecule after the RSEC molecular identifier adjustment algorithm | Sequencing depth |
| Sequencing_Saturation | Percentage of read pairs representing RSEC-adjusted molecules that are sequenced more than once | Sequencing depth |
| Bioproduct_Type | Type of bioproduct in experiment (mRNA or AbSeq) | Assay type |
Cells
| Metric | Definition | Major contributing factors |
|---|---|---|
| Putative_Cell_Count | Number of cell labels detected by the cell label filtering algorithm | Number of cells input and captured by cartridge workflow Bead handling |
| Pct_Reads_from_Putative_Cells | Percentage of reads that are assigned to putative cells | Cell viability Cartridge workflow performance Staining and washing (AbSeq) |
| Mean_Reads_per_Cell | Average number of reads representing the molecules detected in each cell | Sequencing depth |
| Median_Reads_per_Cell | Median number of reads representing the molecules detected in each cell | Sequencing depth |
| Mean_Molecules_per_Cell | Average number of molecules detected per cell label | Sequencing depth |
| Median_Molecules_per_Cell | Median number of molecules detected per cell label | Sequencing depth |
| Mean_Bioproducts_per_Cell | Average number of bioproducts detected per cell label | Sequencing depth |
| Median_Bioproducts_per_Cell | Median number of bioproducts detected per cell label | Sequencing depth |
| Total_Bioproducts_Detected | Number of bioproducts detected from all cells | Sequencing depth |
| Bioproduct_Type | Type of bioproduct in experiment (mRNA or AbSeq) | Assay type |
Sample Tags (if applicable)
| Metric | Definition | Major contributing factors |
|---|---|---|
| Sample_Tag_Filtered_Reads | Number of filtered read pairs aligned to Sample Tags | Sequencing depth |
| ST_Pct_Reads_from_Putative_Cells | Percentage of Sample Tag reads that are assigned to putative cells | Cell viability Sample Tag labeling and wash protocols Cartridge workflow performance Sequencing depth |
VDJ (if applicable)
- See VDJ Metrics output