Pipeline Steps and Algorithms
Introduction
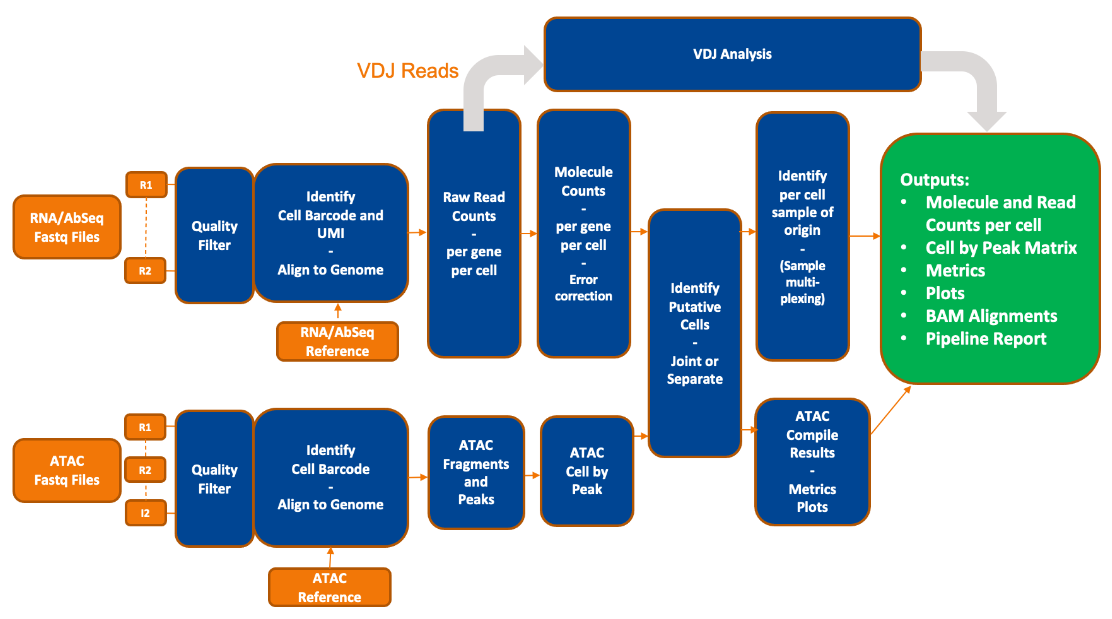
This section provides an in-depth description of each step in the BD Rhapsody™ Sequence Analysis Pipeline. The main portion of these steps decribe the WTA mRNA, Targeted mRNA, AbSeq, and Sample Tag processing steps. For details on the steps of the TCR and BCR analysis or ATAC-Seq analysis, see their dedicated pages.
The BD Rhapsody™ assays are used to create sequencing libraries from single-cell multiomic experiments. For the WTA mRNA, Targeted mRNA, AbSeq, Sample Tag, TCR and BCR libraries, the analysis pipeline works with paired-end FASTQ R1 and R2 files. R1 reads contain information on the cell label and molecular identifier, and R2 reads contain information on the bioproduct. For ATAC-seq libraries the cell label information is on the I2 read, therefore the pipeline additionally requires the I2 FASTQ. The R1 and R2 reads contain information on the genomic DNA fragment. Please refer to the figures below.
Structure of reads generated by sequencing libraries prepared using either the BD Rhapsody™ WTA mRNA, Targeted mRNA,
AbSeq or Sample Tag Assay

Structure of reads generated by sequencing libraries prepared using the BD Rhapsody™ TCR/BCR Full Length Assay

Structure of reads generated by sequencing libraries prepared using the BD Rhapsody™ ATAC-Seq Assay

Note: In the preceding diagrams, CC is an abbreviation for Custom Capture.
Pipeline overview
After sequencing, the pipeline takes input from FASTQ files, a reference (Targeted panel or WTA / WTA+ATAC-Seq reference archive), an AbSeq reference (if required), and a supplemental reference (if required), using these to generate output files and metrics about the pipeline run.