Create Task and Run the Pipeline
Procedure
-
Click the Apps tab to view the apps.
Note: If the app is highlighted in yellow, an update is available. Select the update link to get the latest app version.
-
By the BD Rhapsody™ Sequence Analysis Pipeline, click the green play button under Actions. The Task Inputs tab will display the Inputs and App Settings.
Sequence Analysis Pipeline interface:
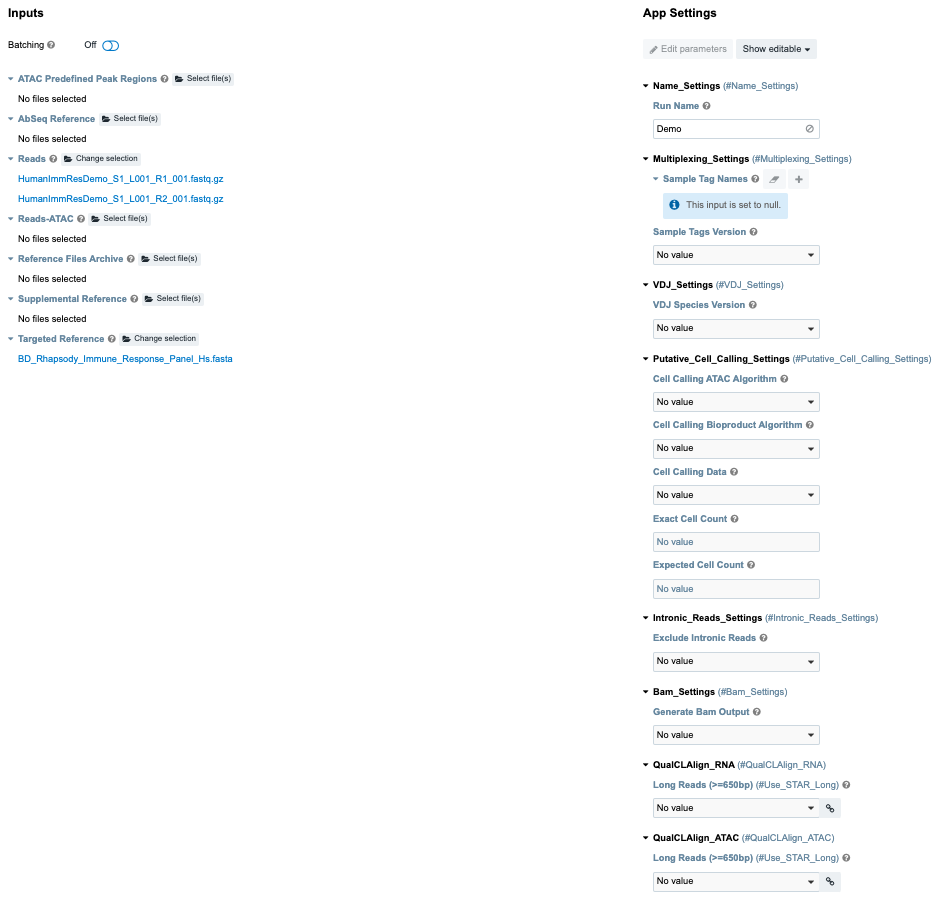
Complete all required fields (which appear in red), and all desired optional App Settings (may depend on assay).
-
In the Inputs section, import your files for analysis according to these requirements:
-
For every
R1.fastq.gzfile, import the pairedR2.fastq.gzfile. For ATAC-Seq, also include the corresponding index readI2.fastq.gzfile. -
Multiple sequencing libraries can be run together as long as they are from the same set of cells, but the files and libraries can be generated from different sequencer runs.
-
Specify at least one reference for the assay type(s) and species of cells in the experiment.
-
-
If necessary, set other input parameters in the App Settings section. For example:
When using a BD® Single-Cell Multiplexing Kit, be sure to select the Sample_Tags_Version (Single-Cell Multiplex Kit
-
Human, Mouse, or Flex) from the dropdown menu.
See Pipeline parameters for details on each app setting.
- Click Run. Seven Bridges Genomics displays the app running on the Tasks tab.
- If you enabled email notifications, look for notification of the completed run.