Molecules and Error Correction
Collapse reads into raw molecules
Reads with the same cell label, same UMI sequence, and same bioproduct are collapsed into a single raw molecule. The number of reads associated with each raw molecule is reported as the raw adjusted sequencing depth.
Remove artifact molecules using RSEC and DBEC UMI adjustment algorithms
PCR and sequencing often generate errors. If the error occurs within the UMI sequence, the R1/R2 read pair is called a unique molecule but is, in fact, an artifact. Artifact molecules contribute to an over-estimated molecule count of a gene in a cell. As sequencing depth increases, the number of raw molecules rises and never plateaus due to these artificial molecules.
To remove the effect of UMI errors on molecule counting, BD Biosciences has developed a set of UMI adjustment algorithms. UMI errors that are single base substitution errors are identified and adjusted to the parent UMI barcode using recursive substitution error correction (RSEC). For targeted sequencing analysis, other UMI errors derived from library preparation steps or sequencing base deletions are later adjusted using distribution-based error correction (DBEC).
Note that targeted sequencing analysis uses RSEC and DBEC, while WTA sequencing analysis uses RSEC only for mRNA libraries and RSEC and DBEC for AbSeq libraries.
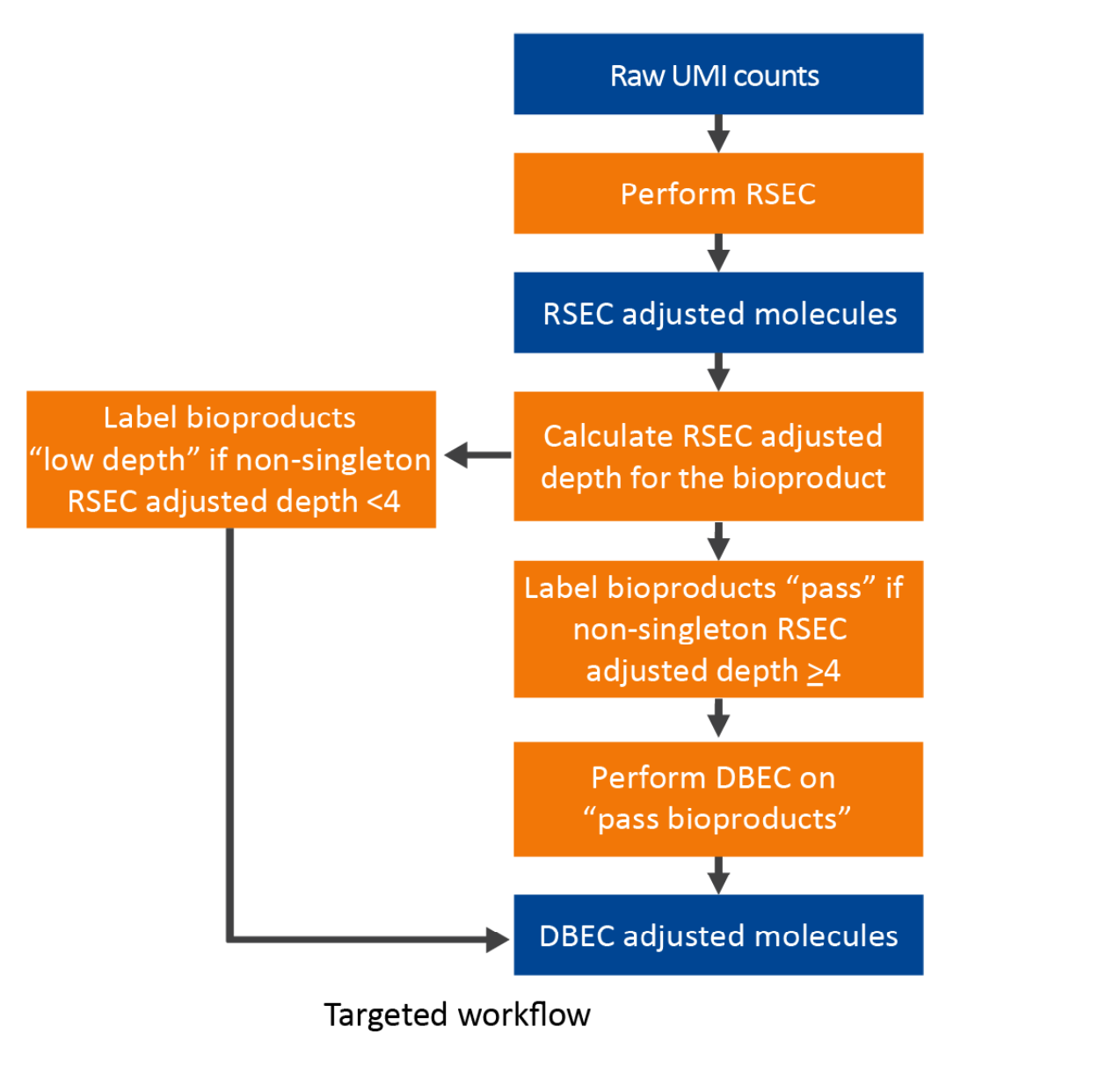
Targeted assay workflow of UMI count adjustment -- using RSEC and DBEC algorithms on both mRNA and AbSeq:
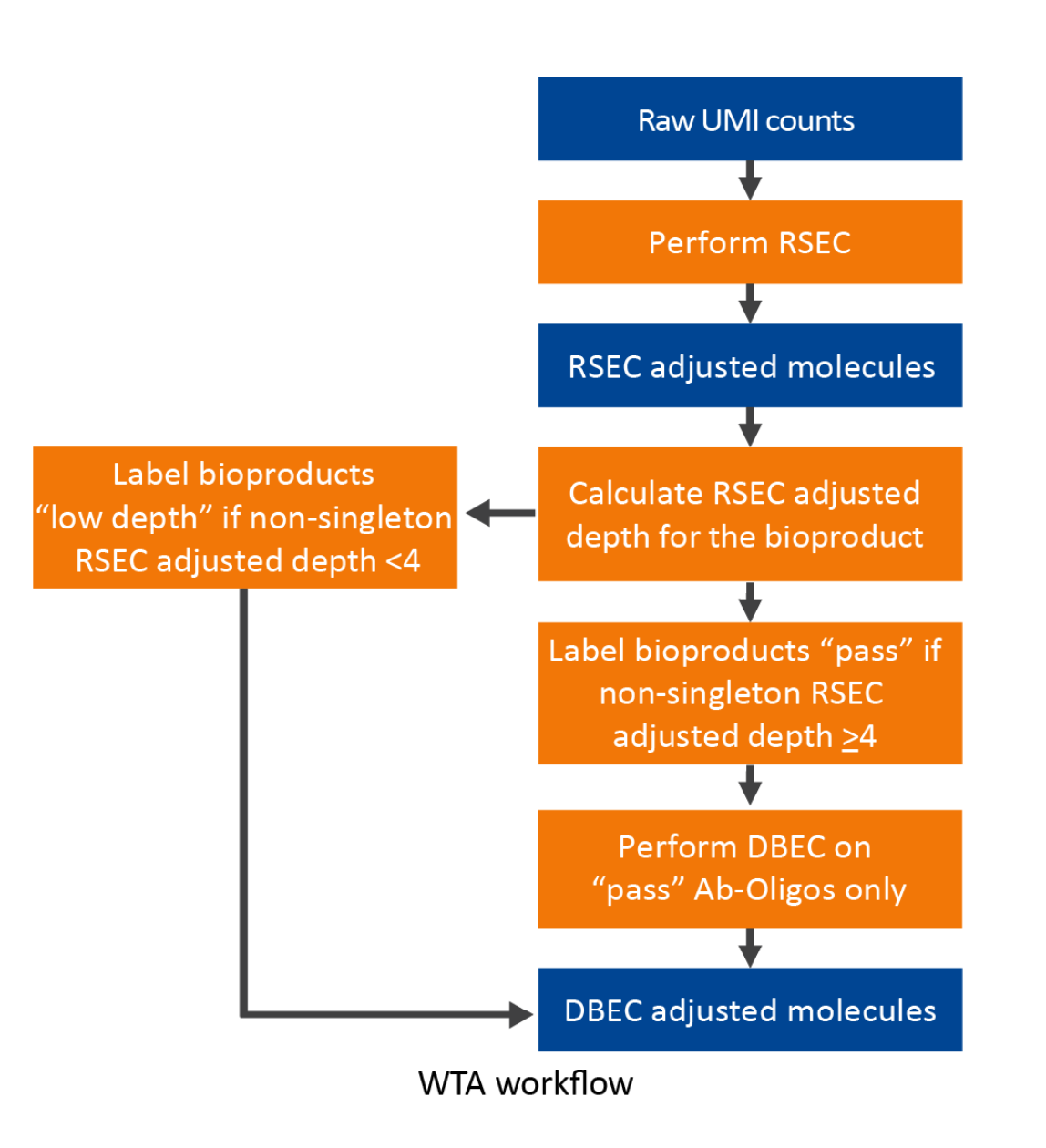
WTA assay workflow of UMI count adjustment -- using RSEC on mRNA, and RSEC and DBEC algorithms on AbSeq:
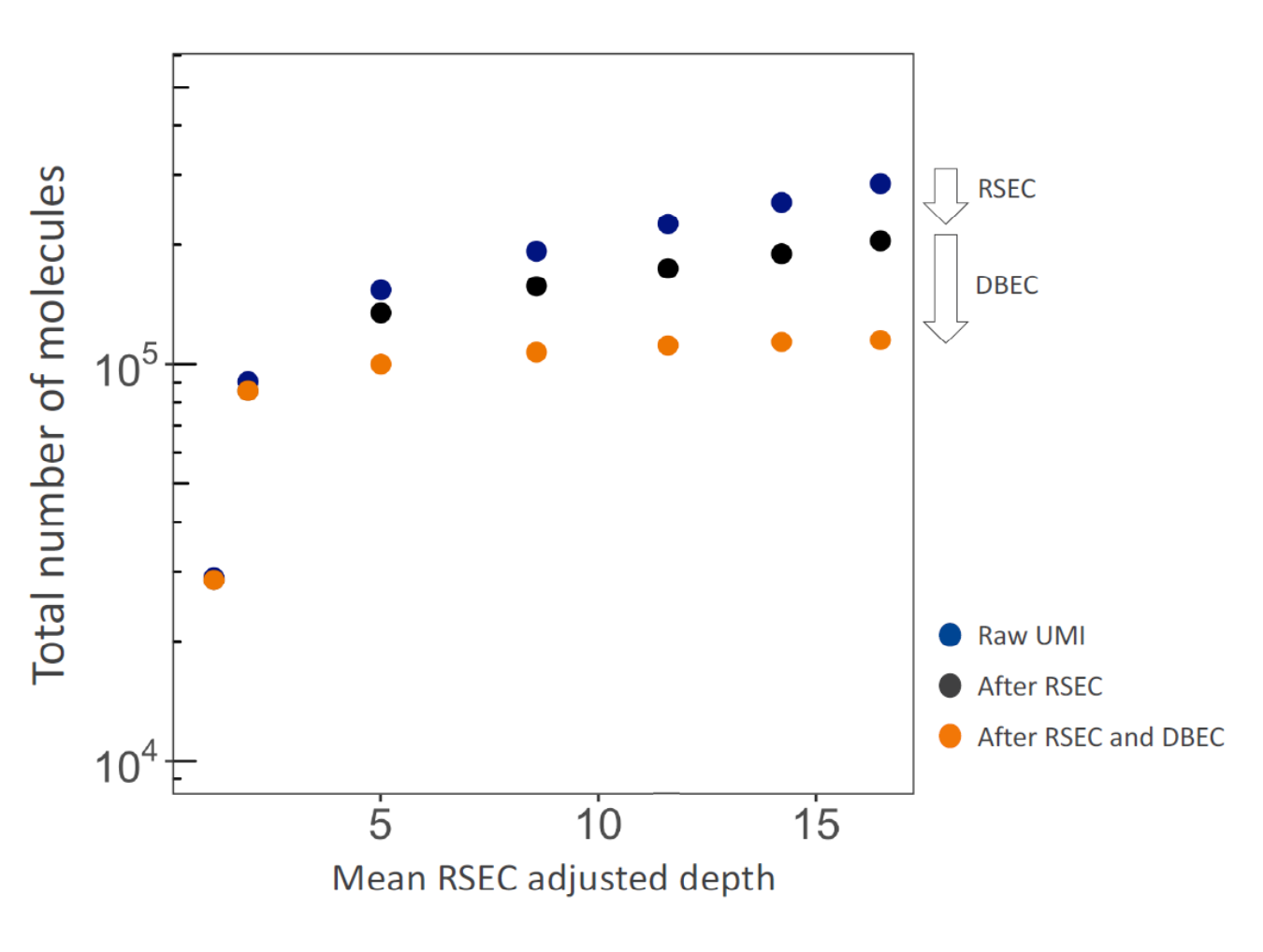
The below graph shows the impact on total molecule count of applying RSEC and DBEC to an example dataset. For targeted
sequencing analysis, if we consider only raw UMIs, the apparent total number of molecules continues to rise with
sequencing depth, because the presence of sequencing and PCR errors contribute to unique UMIs. RSEC removes artifact
molecules from single base substitutions in the UMI sequence. Further adjustment by DBEC removes artifact molecules
originating from PCR errors. As a result, the number of molecules stabilizes with additional sequencing, indicating the
library is sequenced to saturation.
RSEC algorithm to collapse molecules that differ by one base in the UMI sequence
RSEC considers two factors in error correction: 1) similarity in UMI sequence and 2) raw UMI coverage or depth. Below is a somewhat extended example of the recursive error correction algorithm in practice, wherein nine raw UMIs are collapsed into two corrected UMIs.
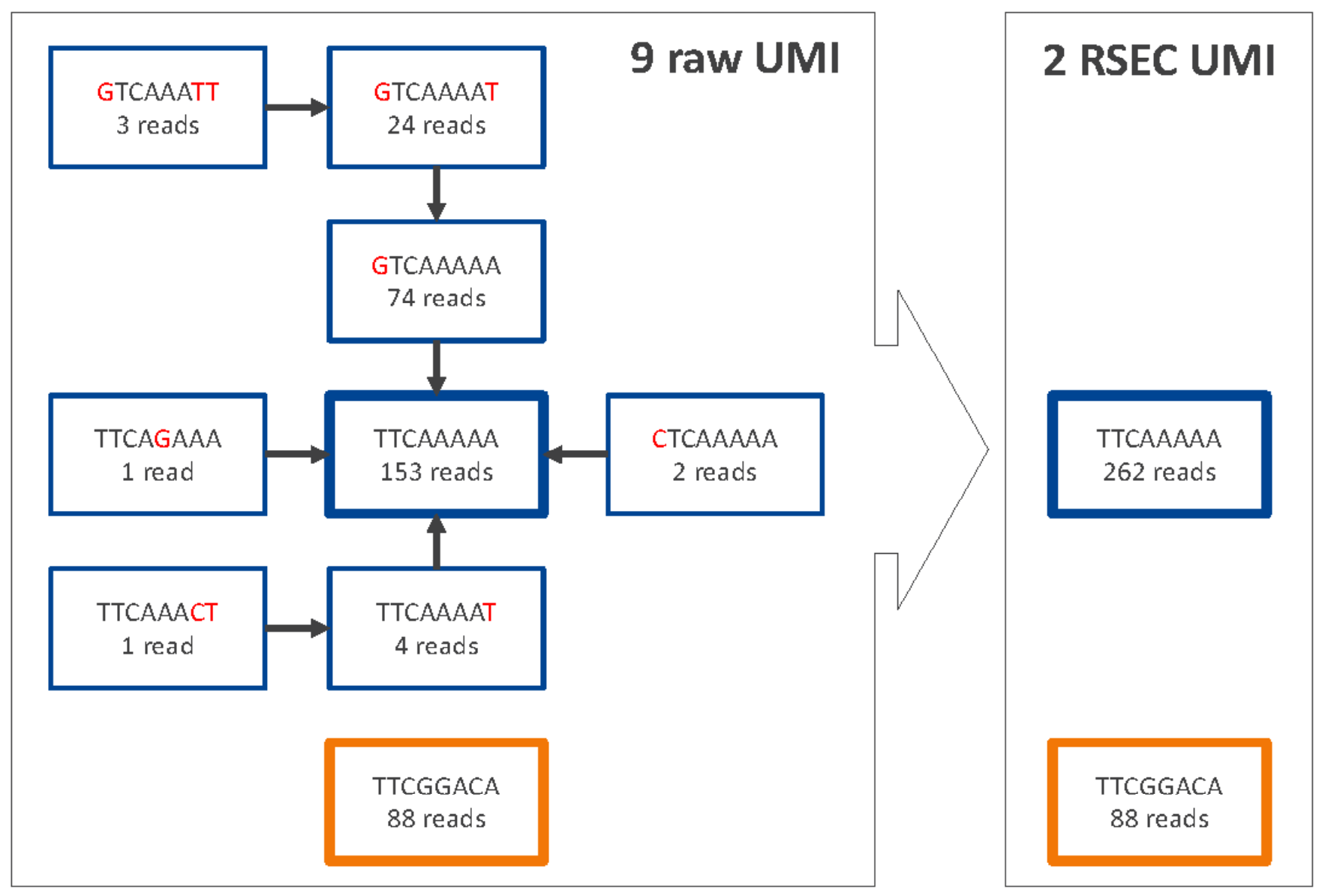
For the molecules from each combination of cell label and bioproduct, UMIs are connected when their UMI sequences are matched to within one base (Hamming distance = 1). For each connection between UMI x and y, if Coverage(y)>2 *Coverage(x)– 1, then y is the Parent UMI and x is the Child UMI. Based on this assignment, child UMIs are collapsed to their parent UMI. This process is recursive until there are no more identifiable parent-child UMIs for the bioproduct.
The number of reads for each child UMI is added to the parent, so no reads are lost. The sum of the reads is the RSEC-adjusted depth of the RSEC-adjusted molecule.
DBEC algorithm to further adjust molecule counts by bioproduct
The RSEC-adjusted molecule counts are further corrected by DBEC, depending on assay type. For Targeted assays, DBEC is applied on all bioproduct types (mRNA and AbSeq). For WTA assays, DBEC is applied only to AbSeq targets.
DBEC is applied on a per-bioproduct basis. The algorithm is based on the assumption that the targeted PCR amplified set of molecules of the same bioproduct, regardless of the cell of origin, is subject to the same amplification efficiency, and therefore, should have similar read depth. Artifact molecules created later in the PCR cycles, such as those derived from PCR chimera formation, will likely have less read depth.
DBEC considers the distribution of RSEC-adjusted depth distribution, not UMI sequence. The sequencing depth of RSEC-adjusted molecules for each bioproduct is a bimodal distribution. The lower mode of the distribution likely represents artifact molecules, and the upper mode likely represents true molecules. The algorithm fits two negative binomial distributions to statistically distinguish between the two modes. Molecules in the upper mode are retained (DBEC-adjusted molecules), while the molecules in the lower mode are discarded. The average depth of the molecules in the upper mode is known as the DBEC-adjusted seq depth. The cutoff between the two modes is the DBEC minimum depth.
See example figure below for gene CCl2. Counts under the orange bars are kept and labeled as DBEC-adjusted molecules. Counts under the blue bars are labeled as erroneous molecules and are discarded. The error depth and DBEC-adjusted depth arrows point to the respective average depths.
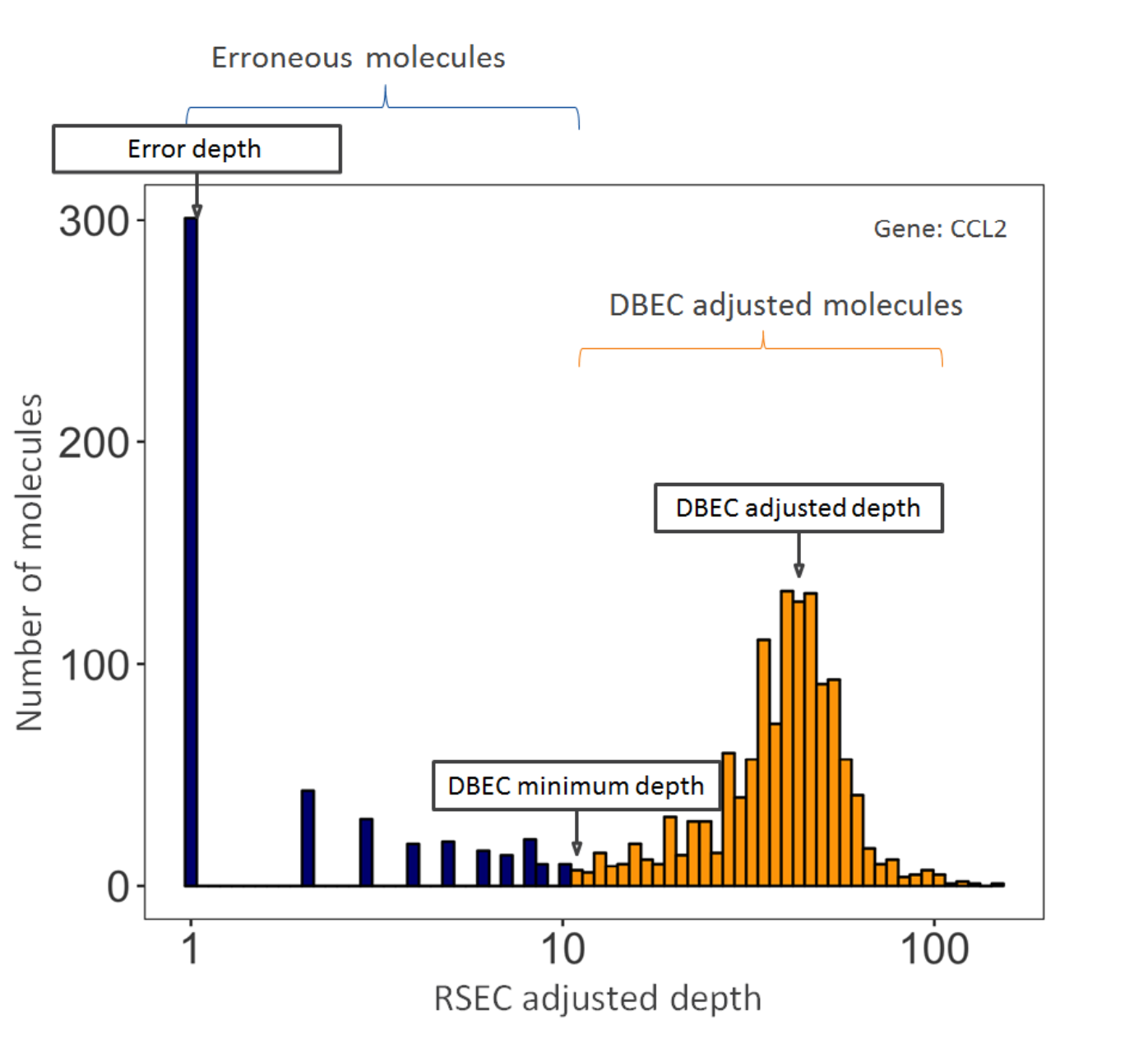
DBEC is applied to bioproducts with an average non-singleton RSEC sequencing depth ≥4. This means that the depth is calculated after removing RSEC UMIs with only one representative read. According to the Poisson distribution, if the average UMI depth is <4, more signal UMIs are removed than error UMIs. As a result, a bioproduct is marked as pass if its average RSEC depth ≥4 and is subject to DBEC. Otherwise, it is marked low depth and bypasses DBEC. If no count is associated with the bioproduct, it is labeled as not detected.
DBEC removes molecules and the reads associated with the removed molecules from consideration in downstream analyses.
The RSEC and DBEC metrics associated with each bioproduct are reported in the file,
<sample_name>_Bioproduct_Stats.csv.